New IRTG 2403 Projects starting 2022
Prof. Dr. Kerstin Kaufmann (Humboldt-Universität zu Berlin)
Please contact Prof. Dr. Kerstin Kaufmann directly for possible projects.
https://www2.hu-berlin.de/biologie/flower/
Prof. Dr. Stefan Mundlos (Max Planck Institute for Molecular Genetics)
Genomes in Evolution – How Bats learned to fly
Evolutionary processes have resulted in the most diverse adaptions to environmental challenges. How such adaptations evolve and how they are encoded in the genome remains one of the great mysteries in biology. Here, we follow the hypothesis that a high degree of the morphological diversity in the animal kingdom can be attributed to changes in the regulatory genome and that these changes can be identified using recent technological advances including the generation of high-resolution genomes, epigenetic mapping of regulatory sequences and single-cell sequencing. In a previous study we successfully applied this strategy to investigate the genomic origin of intersexuality in female moles (Real et al. 2020).
Here, we want to elucidate a fascinating phenomenon in evolution, the development of wings in bats. In a preliminary study, we generated a full chromosome genome of the short tailed fruit bat (Carollia perspicillata). Furthermore, we collected fore- and hindlimbs from bat embryos (Carollia perspicillata), covering limb development from the early limb bud to the later stages of skeletal growth. Using these tissues, we produced ChIP-seq and ATAC-seq data and generated a genome-wide map of enhancers. In addition, we generated single-cell RNA-seq (sc-RNA-seq) as well as sc-ATAC-seq data from these tissues. An equivalent data set was generated from mouse embryos. This unique dataset provides us with novel insights into limb development and how it can be modified to generate extreme differences in morphology. In an integrated data analysis we will identify regulated genes in Carollia limb development and changes in their corresponding regulatory landscapes. Identified regulatory regions consiting of enhancers, promoter and/or lncRNAs will be reconstituted in mice by genomic engineering. For this purpose we will synthetically produced large DNA sequences using a yeast assembly protocol and insert them into the mouse genome. Transgenic mice will be used to dissect how genomic changes translate into altered gene expression and phenotypes on cellular and regulatory level. Finally, we will create de novo designer regulatory landscapes that can be used as a testbed for experimental perturbations.
The possibility to re-engineer sequences in another species will provide us with an unprecedented insight how non-coding DNA regulates gene expression and how this translates into phenotype.
M Real F, et al. The mole genome reveals regulatory rearrangements associated with adaptive intersexuality. Science. 2020 Oct 9;370(6513):208-214.
Please contact Prof. Dr. Stefan Mundlos directly for possible projects.
https://www.molgen.mpg.de/Development-and-Disease
Dr. Andreas Mayer (Max Planck Institute for Molecular Genetics)
Revealing BET protein-specific functions in transcription and chromatin regulation
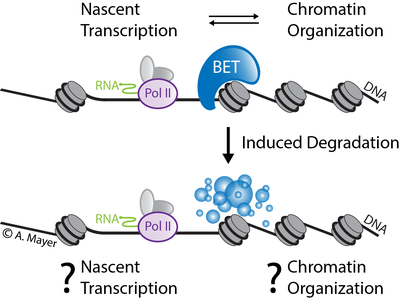
Our knowledge of the regulatory crosstalk between RNA polymerase II (Pol II) transcription and chromatin organization to control cell function is still incomplete. In this project, we will focus on elucidating the specific role of BET bromodomain proteins in transcription and chromatin regulation. BET proteins act at the interface of Pol II transcription and chromatin structure regulation. The most prominent member of the BET protein family BRD4 has emerged as a therapeutic target in a range of human diseases. Here, we will study the functions of BET proteins in different human cellular models and during stem cell differentiation. We will use functional genomics approaches including advanced computational methods (machine learning modeling, multi-omics data integration, visualization) to infer and predict direct functions of BET proteins in the regulatory transcription-chromatin interface. We encourage students with a strong background in computational science (Master’s degree in Bioinformatics or in similar disciplines) to apply. In case of interest please also visit the website of the Mayer Group which is located at the Max Planck Institute for Molecular Genetics.
Prof. Dr. Ana Pombo (Humboldt-Universität zu Berlin)
Please contact Prof. Dr. Ana Pombo directly for possible projects.
https://www.mdc-berlin.de/pombo
Dr. Edda Schulz (Max Planck Institute for Molecular Genetics)
Mapping funktional enhancer-transcription factor interactions
While genome-wide mapping of transcription factor (TF) binding sites is becoming increasingly easy, comprehensive identification of all TFs that regulate a specific enhancer element is not yet possible. To address this challenge we will establish a high-throughput assay for functional mapping of TF-enhancer interactions. To this end we will combine a pooled CRISPR screen with a massively parallel reporter assay (MPRA). Once established, the assay will be applied to precisely map TF binding motifs in enhancer elements of the Xist gene that we have recently identified (Gjaltema, Schwämmle er al, BioRxiv, 2021).
Dr. Laleh Haghverdi (Max-Delbrück-Centrum für Molekulare Medizin (MDC))
Incorporating Gene Regulatory Networks in the Learning Process on High-Throughput Single-Cell Omics Data
The advent of new single-cell high-throughput technologies has opened up great opportunities for application of powerful deep learning approaches for extraction of accurate biological information from large amount of available data. Nevertheless, existing application of deep learning for single-cell omics analysis has mostly overlooked a property in such data, namely the notion of modularity of the underlying gene regulatory networks (GRNs).
Modular data structures are immediately present in other popular deep learning applications including image, audio and speech data types with proven advantages for more efficient learning models such as convolutional neural networks (CNNs) in the case of regular, modular graphs (i.e., images). In this project, we explore geometrical deep learning approaches for incorporation of the modular structure of GRNs in learning processes on high-throughput single-cell omic datasets.
Current machine learning practices in the field of learning on omics data, treat genes as independent features. Yet, we are aware that genes as well are not single and unrelated features; they interact with each other through the cell’s regulatory circuitry and in fact, most cellular processes are carried out through modules of genes rather than single gene’s function [Ravisi et al. 2010, Davidson 2010]. A few studies have used scRNA-seq data to infer such modular gene relations [Farrell et al. 2018, Wagner et al. 2016]. There are also a growing number and progression of databases [Van de Sande et al. 2020, Holland et al. 2020] that incorporate validated regulatory relations between the transcription factors and downstream genes for inferring the underlying GRN in a data set. We propose that using such prior knowledge of GRNs, can benefit our learning capabilities and outcomes and by imposing a biologically meaningful inductive bias.
In the first phase of the project (in progress) we design and apply a geometrical deep learning approach for learning on GRNs as irregular but modular graphs that are validated by public database for cell type classification using transcriptomics data. In the second phase, we will adopt this approach for robust inference of lineage trajectories in cell differentiation. Furthermore, we will explore new directions for extensions of our framework for application on other omic data modalities such as epigenomics and proteomics.
Dr. Martin Kircher (Charité)
Modelling regulatory sequences and tissue-specificity using functional read-outs
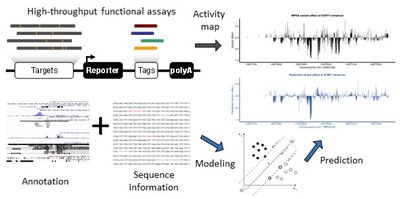 Understanding how gene regulation is encoded across development and the diversity of cell-types remains a fundamental challenge in biological research. While large-scale experiments and coordinated efforts have provided us with an unprecedented amount of experimental data, our current understanding of how gene regulation is encoded on the genomic level is still limited. The Kircher lab devises computational approaches (e.g. CADD, CADD-SV, ReMM) to score and identify functionally relevant genetic changes in the human genome. In collaboration with others, we use activity measures from reporter assays (MPRA/CRE-Seq) and CRISPR/Cas screens of regulatory sequences (i.e. enhancer and promoter elements) in combination with additional functional read outs (like chromatin accessibility and histone marks) from the same cell-type and/or condition to study expression effects. Our collaborators include for example the labs of Gregory Crawford at the Duke Center for Genomic and Computational Biology, Jay Shendure at University of Washington Seattle, and Nadav Ahituv at the University of California San Francisco.
Understanding how gene regulation is encoded across development and the diversity of cell-types remains a fundamental challenge in biological research. While large-scale experiments and coordinated efforts have provided us with an unprecedented amount of experimental data, our current understanding of how gene regulation is encoded on the genomic level is still limited. The Kircher lab devises computational approaches (e.g. CADD, CADD-SV, ReMM) to score and identify functionally relevant genetic changes in the human genome. In collaboration with others, we use activity measures from reporter assays (MPRA/CRE-Seq) and CRISPR/Cas screens of regulatory sequences (i.e. enhancer and promoter elements) in combination with additional functional read outs (like chromatin accessibility and histone marks) from the same cell-type and/or condition to study expression effects. Our collaborators include for example the labs of Gregory Crawford at the Duke Center for Genomic and Computational Biology, Jay Shendure at University of Washington Seattle, and Nadav Ahituv at the University of California San Francisco.
We model expression effects from sequence in convolutional neural nets or integrate sequence features, DNA shape, epigenetic marks, and interactions between DNA binding factors in classical machine learning models. Our goal is to improve our understanding of regulatory sequences and to develop comprehensive and generalizable models of regulatory effects in the human genome, which can be integrated into predictors of genome-wide variant effects.
Prof. Dr. Uwe Ohler (Humboldt-Universität zu Berlin)
Regulatory grammars from synthetic and single-cell data sets
In our project, we are looking for an experimental and/or computational biology student, who wants to work on the interface between machine learning and synthetic regulatory genomics.
The goal of the project is to use deep learning models to learn regulatory grammars from synthetic and single-cell data sets, and to develop and apply approaches that will allow us to use these models to create new sequences with a specific function: To guide the design of new synthetic training sequences that improve predictive performance, and eventually of new regulatory regions with designed expression patterns in vivo. We aim to pursue this in carefully controlled scenarios and model systems, such as A thaliana in collaboration with the lab of Philip Benfey.
Prof. Dr. Martin Vingron (Max Planck Institute for Molecular Genetics)
Epigenetic enhancers and sequence conservation
Traditionally, enhancers were assumed to show sequence conservation because their regulatory role was important and the sequence therefore under selection pressure. This is particularly obvious for many developmental enhancers. Today, enhancers are often characterized in a cell-type specific manner using epigenetic marks, without considering sequence conservation. In this project we want to search for traces of selective pressure on these epigenetically defined enhancers, and hopefully characterize which enhancers are conserved in sequence. (An entry point into the literature on this question is http://www.genome.org/cgi/doi/10.1101/gr.203679.115 )
